CELL BIOLOGY
Overview of Cell Biology:
Prokaryotic cells of the Eubacteria and Archaea differ from eukaryotic cells in genetics, metabolism, and cell structure (lack of a nuclear membrane, lack of organelles, differences in cell walls). Prokaryotic flagellae and adhesion molecules participate in prokaryotic chemotaxis.
Table Comparisons of Eubacteria, Archaea, and Eukaryotes Cell walls of Prokaryotes Electron acceptors for respiration and methanogenesis in prokaryotes Glycolysis in bacteria Lithotrophic prokaryotes Comparison of Plant and Bacterial Photosynthesis Structure of bacteriochlorophylls
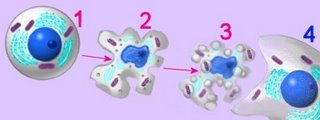
The cells of eukaryotic organisms are larger and structurally more complex than those of prokaryotes. Eukaryotic cells are divided into functional compartments (cytoplasm, organellar lumens, nucleus, vacuoles, vesicles) by a variety of cell membranes. Membrane-bound subcellular energy plastids (chloroplasts, mitochondria) were acquired through serial endosymbiotic events about 1 billion years ago. Cellular compartments include the nucleus, which communicates with the cytoplasm by way of nuclear pores and the endoplasmic reticulum. Cytoplasmic compartments include the cytoskeleton (microtubules, microfilaments, intermediate filaments, basal bodies and centrioles) specialized vesicular structures such as the endoplasmic reticulum, Golgi apparatus, proteasomes, lysosomes, plant vacuoles, endosomes and exosomes, and energy organelles (chloroplasts, mitochondria).
Cells perform a wide variety of physiological functions such as active transport and intracellular transport; proliferation, differentiation, the cellular stress response, and programmed cell death; they respond to the environment through chemotaxis, energy transduction, and signaling; and move materials into the cell through phagocytosis, pinocytosis, and receptor-mediated endocytosis.
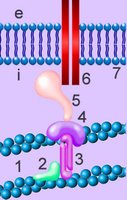
Cell membranes comprise phospholipids, sugars, and specialized proteins and perform a variety of functions such as adhesion, cellular communication, and maintenance of concentration gradients by active transport. These activities procede by means of specialized adhesion molecules, signaling molecules, ion channels and pumps, and receptor proteins.
The cell's cytoskeleton participates in chemotaxis, intracellular transport, and cell-to-cell adhesion. The cytoskeleton interacts with the extracellular environnment by way of transmembrane adhesion molecules (CAMs) and through surface protuberances such as cilia and flagellae (with various functions as mechanoreceptors, chemoreceptors, and the outer segment of the rods in the vertebrate retina). The centriolar components of the cytoskeleton organize the spindle apparatus on which the nuclear chromosomes translocate during mitosis and meiosis, phases of the reproductive cell cycle.
In multicellular organism, proliferation of cells must be balanced by cell death, usually through programmed, non-inflammatory apoptosis.
Cell Adhesion Molecules Immune Cytokines Second Messengers Cell signaling
Apoptosis vs Necrosis Apoptosis
This site deals with Cell Biology, Cell Signaling, and the Molecular Biology of cells, particularly in relation to genetic mechanisms of biological evolution. Blue terms hyperlink to explanatory items so that you may navigate through items providing more detail. The site is searchable using the "Search this blog" box at top left. Use the "back" function to return to each departure item.
Alphabetic listing of Items - main items are bold
CELL BIOLOGY : A : active transport : adhesion : apoptosis : C : cell cycle : cell membranes : centrioles : (chemical gradients) : cilia and flagella : communication : concentration gradients : cytoplasm : cytoskeleton : D : death of cells : E : endocytosis : endoplasmic reticulum : endosomes : energy transducers : eukaryotic : exosome : F : flagella : G : Golgi apparatus : I : ion channels : L : lysosome : M : meiosis : microtubules : mitochondrion : mitosis : mitotic spindle : N : nuclear membrane : nuclear pore : nucleolus : nucleus : P : peroxisome : transport : phagocytosis : photosynthesis : physiological function : pinocytosis : plant cell : plasma membrane : prokaryotic : proteasome : protein degradation : protein pumps pumps : R : receptor proteins : receptor-mediated endocytosis : reproduction : ribosomes : RTKs : S : spindle : structure : V : vacuole : vesicle :
CELL SIGNALING : AKAPs : cellular signal transduction : concentration gradients : chemical gradients & communication : chemotaxis : cytokines : cytokine receptors : DAG : DAGKs : diacylglycerol : diacyl glycerol kinase : ERKs : GPCRs : GPCR families : hormones : neural action neural activity neuronal activity neuronal interconnections : neurotransmission : neurotransmitters : Nitric Oxide : PDZ domain : phosphorylation : phosphotransfer-mediated signaling pathways : PKA, protein kinase A : phospholipase C-gamma : PLC-G : PKC : protein kinase A : protein kinase C : protein tyrosine kinases protein kinases : Protein Kinase Signaling Networks : Ras : second messengers : signaling gradients (concentration gradients : chemical gradients) : signal transduction : two-component systems :
MOLECULAR BIOLOGY : amino acids : caspases : (concentration gradients) : (hormones) : proteins : Chemistry of Life site Biochemistry & Molecular Genetics : Molecular Genetics site
Items occur within Sections (listed in the sidebar). Items are listed in groups of 10 – to see more items, click on the lowest item in the sidebar. When visiting an item, the site title changes to purple – click on the title or “Home” to return to the main page.
The “Guide-Glossary” link below each item provides a glossary of terms ( # > 0 ), as a pop-up when reading within a Section, or as sub-script when visiting an Item. | 0 Guide-Glossary